TARGETED THERAPEUTICS: AN INTRODUCTION
by Eric P. Hoffman PhD and John McCall PhD
Source: Fall/Winter 2006 CCCF Newsletter
There are many pharmaceutical drugs on the market, and many of them work quite well. Unfortunately, many kids with cancer are not helped by those that are available, and the cancer goes on relatively unchecked (see accompanying article “ACCOs Expands Mission“).
How are drugs “discovered,” and how can we discover new drugs for kids with cancer? This article reviews the general protocol used by pharmaceutical companies to discover new drugs known as targeted therapeutics, or “high throughput screening” (HTS). We then discuss how there are aggressive efforts to move parts of the HTS pipeline from pharmaceutical companies to the academic medicine (public) research domain. We discuss key recent developments in genomics that are speeding up the initial step of HTS, namely “target identification.” Finally, we discuss the costs associated with HTS for specific disorders, and provide examples of how costs are being shared between different sources (government, patient organizations, foundations, and others).
Looking for Help?
ACCO offers FREE Books & Resources for Families of Children with Cancer
TARGETED THERAPEUTICS 101 AND THE HIGH THROUGHPUT SCREENING (HTS) PIPELINE
What does “targeted therapeutics” and “HTS pipeline” mean? Everyone is made of cells; small living self-contained building blocks, where each microscopic cell in blood, bone, and other tissues contain the same set of instructions. The instructions for telling a cell how to behave is all contained in the DNA – the comprehensive cookbook containing all 30,000 recipes needed to construct a human from conception through old age. The bone cells read the recipes (genes) to tell them how to behave as a bone cell, and the blood cells read the genes to act appropriately for blood. It is important to emphasize that each cell contains the same comprehensive cookbook, but reads only the appropriate parts at the appropriate times. In reading the genes/recipes, the cell makes proteins that all interact with each other, much like an assembly line; a bone cell makes all the proteins needed to produce hard calcified material (like the assembly line for the steel girders of a office tower), while the nerve cells are reading the instructions for the assembly line of a desk top computer, and blood cells reading genes for the assembly line for a plumbing system.
Cancer results when cells start reading the wrong genes at the wrong time or place; much like a nerve cell inappropriately reading the bone instructions for steel girder assembly. Building a lot of bone inside your brain can not lead to much good. Leukemia often result from a blood cell reading instructions for “rapid reproduction,” leading to the cell making billions of offspring (cell babies), when this is entirely inappropriate for this specific blood cell.
The goal of targeted therapeutics is to understand precisely what inappropriate recipes (genes) are being read by the specific cancer, then getting drugs to knock out that specific inappropriate assembly line (pathway). For example, using the analogy of a nerve cell inappropriately carrying out bone cell instructions to build steel girders, scientists go into the molecular pathways in the cancer, and look for the initial step to block it. In this analogy, scientists discover that the first incorrect instruction is “deliver a few truckloads of steel beams to the door of the nerve cell so that it can start the assembly line for building steel framework.” If a drug can be found to stop the truck delivery, then the nerve cell can not even start making the steel girder assemblies – it is stopped from acting inappropriately by blocking the first mis-step.
The oft-touted Gleevec is a good example of targeted therapeutics, or “molecular designer drug.” The first mis-step by cells in chronic myelogenous leukemia(CML) is often a fusion of two recipes (Philadelphia chromosome translocation). A blood cell gets two recipes confused, so that a signaling protein (tyrosine kinase) is turned on in the blood cells when it shouldn’t be. The tyrosine kinase tells the blood cells to start multiplying rapidly, and to not respond appropriately to instructions from neighboring blood cells to slow down. Given that this specific tyrosine kinase drives much of the CML cancer, scientists realized that if they could knock out this inappropriate tyrosine kinase signal, then the cancer would be stopped at the first step. This was true, and now the majority of patients with CML are treated and remain cancer free by using Gleevec.
Targeted therapeutics then is to identify the key early events in inappropriate behavior by cancers, and develop drugs specifically targeted to the recipes/ genes causing these early mis-steps of the cell.
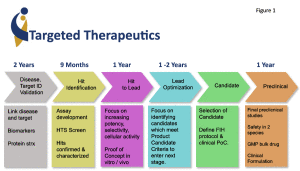
How does one develop targeted therapeutics for childhood cancer? There are a number of steps in the “pipeline” (Figure 1), with the first two being “target identification” (target ID), and the second development of experimental systems to identify drugs that knockout that specific target (high throughput drug screening; HTS).
Target ID involves characterizing most if not all of the recipes (genes) being read by the cancer cell, to see how these differ from what that cell should normally be reading and doing. The method used for seeing which of the 30,000 recipes are being read by the cancer cell is called “gene expression profiling,” and uses microarrays of all genes arrayed on small glass slides to monitor use of each gene in the genome. A second method, comparative genomic hybridization (CGH) is used to determine if any of the instructions (30,000 genes in DNA) are missing or amplified (too many copies of the gene) in the tumor cells. A third approach is to read genes letter-by-letter, looking for very subtle changes in the recipe that could cause inappropriate behavior, thus leading to cancer. For example, reading a cake recipe that calls for “salt,” but changing one letter to “silt” would lead to a lot of river dirt being added to your cake. Alternatively, most or all of the proteins (actual pathways or assembly lines) can be tested by a method called “proteomics.” Studying all of these DNA, mRNA, and protein components of a wayward cancer cell, and seeing which are different from its normal cell counterparts, leads to a list of “targets” that stand a good chance of stopping the cancer.
Once “targets” are in hand, the next step is to find drugs that knockout one or more of those targets to whip the cancer cell back in shape (back to a normally behaving cell). This approach is considerably different from standard cancer chemotherapy. Chemotherapy aims to kill cancer cells, with considerable “collateral damage” of normal cells in the process. Targeted therapeutics aims to do “behavior modification” of cancer cells, rather than necessarily kill them.
How does one identify a “targeted drug?” This is typically done using high throughput drug screening, or HTS. A model of the “target” is designed so that cells in a Petri dish are able to tell the researcher when a drug is modifying the target. Many of these “cell based assays” involve the cell shining light when the desired behavior modification is achieved. The researcher can then test thousands of drugs, to look for the few drugs that make the cell “light up,” letting the researcher know that that specific drug knocked out the target.
Drug “hits” are then further tested for a variety of features to make them “good drugs” that can be given to a patient without serious side effects, but are effective against the cancer. This always includes clinical trials to prove that the drug works in patients as predicted (Figure 1).
WHO DOES THE TARGETED THERAPEUTICS PIPELINE?
Pharmaceutical and biotechnology companies have become experts at the targeted therapeutics HTS pipeline over the last 20 years. Drug companies develop projects on specific diseases and targets; examples include protease inhibitors in AIDS, cox-2 inhibitors (Vioxx and Celebrex in pain), and many others. The process is expensive, and there is a lot of attrition at each step of the HTS pipeline. In order to develop one good targeted drug “on market,” the pharmaceuticals calculate on pursuing over 50 initial targets. It is also an expensive process, with calculations hovering around $500 – $800 million spent per targeted drug that ends up receiving marketing approval by the Federal Drug Agency (FDA).
Because of the cost, pharmaceuticals tend to focus on only the most common disorders that have the most potential for future income from sales of the drug (so-called “blockbuster drugs”). Fortunately, childhood cancer is relatively rare. However, this has the unfortunate corollary that pharmaceutical companies are generally not interested in pursuing targeted therapeutics for childhood cancer.
This problem of lack of attention by major pharmaceuticals is not unique to childhood cancers. Children are generally healthier than older people, so childhood diseases are relatively rare, with relatively little “market potential.” As a result, there are very few pharmaceutical programs for targeted therapeutics in any childhood disease, as well as scores of less common adult disorders.
The government research portfolio, National Institute of Health (NIH), National Cancer Institute (NCI), and increasingly the Department of Defense (DOD), often attempts to compensate (or complement) the money-oriented pharmaceutical research and business machine with academic research and clinical trials in university medical centers. With the appointment of the new Director of the NIH, Elias Zerhouni, there has been a recent yet very strong push to have academic medical centers pick up some of the responsibility for the targeted therapeutics and HTS pipeline. Over the last few years, an impressive number of new funding and programs have sprung up to meet this challenge. Indeed, many universities are starting collaborative groups and divisions that appear much like small pharmaceutical companies.
It is still too early to judge the effectiveness of these new government programs, as new drugs take a few years to develop (Figure 1). Hopefully, these academic efforts should lead to scores of new targeted drugs for childhood cancer and other diseases, and a new class of public/private partnerships as more and more of the “early stages” of targeted therapeutics research is done by the public academic centers.
GENOMICS, PROTEOMICS AND PUBLIC ACCESS:
THE ACADEMIC KEY TO UNLOCKING TARGETED THERAPEUTICS FOR CHILDHOOD CANCER
There are three inter-related aspects of the “academic HTS pipeline” that together, may result in a “sea change,” and a dramatic surge forward in targeted drug development: 1/ parallelization of research, 2/ emerging technologies, and 3/ public access. At any point in time, a major pharmaceutical may be focused on only a handful (7-10) targets in their drug development programs. This is a woefully small number, considering both the need (different disorders, with dozens of types of childhood cancer alone), and the attrition. By opening target identification up to the academic sector, thousands of laboratories could begin functioning as miniature drug companies, with parallelization of each step of the HTS process in a global university network. Emerging technologies such as microarrays and proteomics are becoming robust approaches to define the molecular fingerprint of each tumor type, and in many respects the academic sector is more accomplished at “target ID” than major pharmaceuticals.
But perhaps most importantly is the strong push to quickly move all data generated by academic centers into the public domain. Pharmaceutical companies rarely if ever release its research data to the public. This is considered proprietary research data for their own drug discovery programs, and is held close “to their chest.” In contrast, NIH is developing increasing numbers of programs that are requiring government-funded researchers to release massive amounts of microarray, CGH, proteomic, and sequencing data into publicly accessible databases, often prior to any formal publication of the data. Thus, microarray data generated on medulloblastoma brain tumors at Children’s National Medical Center in Washington DC for example, can be analyzed by scientists in China to identify new potential drug targets, and thus propel the pace of research dramatically forward.
As discussed elsewhere in this newsletter, it is imperative to begin Target ID and HTS screening programs for each of the pediatric cancers. Once started, these programs will be heavily subsidized by new NIH and NCI programs, and will become highly parallelized through the global research network.